3. Inferring iModulon activities for new data
Re-computing the complete set of iModulons can be computationally intensive for every new dataset. However, once a dataset reaches a critical size, you can use a pre-computed IcaData object to infer the iModulon activities of a new dataset. iModulon activities are relative measures; every dataset must have a reference condition to which all other samples are compared against.
To compute the new iModulon activities, first load the pre-computed IcaData object.
[1]:
from pymodulon.example_data import load_ecoli_data
ica_data = load_ecoli_data()
Next, load your expression profiles. This should be normalized using whichever read mapping pipeline you use, as Transcripts per Million (TPM) or log-TPM.
[2]:
from pymodulon.example_data import load_example_log_tpm
log_tpm = load_example_log_tpm()
log_tpm.head()
[2]:
| Reference_1 | Reference_2 | Test_1 | Test_2 | |
|---|---|---|---|---|
| Geneid | ||||
| b0001 | 10.473721 | 10.271944 | 10.315476 | 10.808135 |
| b0002 | 10.260569 | 10.368555 | 10.735874 | 10.726916 |
| b0003 | 9.920277 | 10.044224 | 10.528432 | 10.503092 |
| b0004 | 9.936694 | 10.010638 | 9.739519 | 9.722997 |
| b0005 | 7.027515 | 7.237449 | 6.745798 | 6.497823 |
Next, make sure your dataset uses similar gene names as the target IcaData object.
[3]:
from matplotlib_venn import venn2
venn2((set(ica_data.gene_names),set(log_tpm.index)), set_labels=['IcaData genes','Dataset genes'])
[3]:
<matplotlib_venn._common.VennDiagram at 0x7f6f9c174650>
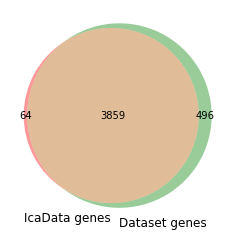
Only genes shared between your IcaData object and the new expression profiles will be used to project your data. All other genes will be ignored.
Then, center your dataset on a reference condition, taking the average of replicates.
[4]:
centered_log_tpm = log_tpm.sub(log_tpm[['Reference_1','Reference_2']].mean(axis=1),axis=0)
centered_log_tpm.head()
[4]:
| Reference_1 | Reference_2 | Test_1 | Test_2 | |
|---|---|---|---|---|
| Geneid | ||||
| b0001 | 0.100889 | -0.100889 | -0.057356 | 0.435303 |
| b0002 | -0.053993 | 0.053993 | 0.421312 | 0.412354 |
| b0003 | -0.061973 | 0.061973 | 0.546181 | 0.520841 |
| b0004 | -0.036972 | 0.036972 | -0.234147 | -0.250669 |
| b0005 | -0.104967 | 0.104967 | -0.386684 | -0.634659 |
Finally, use the pymodulon.util.infer_activities function to infer the relative iModulon activities of your dataset.
[5]:
from pymodulon.util import infer_activities
[6]:
activities = infer_activities(ica_data,centered_log_tpm)
activities.head()
/home/docs/checkouts/readthedocs.org/user_builds/pymodulon/envs/stable/lib/python3.7/site-packages/pymodulon/util.py:327: FutureWarning: Index.__and__ operating as a set operation is deprecated, in the future this will be a logical operation matching Series.__and__. Use index.intersection(other) instead
shared_genes = ica_data.M.index & data.index
[6]:
| Reference_1 | Reference_2 | Test_1 | Test_2 | |
|---|---|---|---|---|
| AllR/AraC/FucR | 0.243143 | -0.243143 | 1.028044 | 0.848571 |
| ArcA-1 | -0.157687 | 0.157687 | -2.644027 | -2.418106 |
| ArcA-2 | 0.038248 | -0.038248 | 0.182260 | 0.039267 |
| ArgR | -0.150147 | 0.150147 | -1.456806 | -1.293399 |
| AtoC | 0.344893 | -0.344893 | 0.632130 | 1.075412 |
All of the plotting functions in pymodulon.plotting can be used on your inferred activities once you add it to a new IcaData object. It is advisable to create a new sample_table with project and condition columns.
[7]:
from pymodulon.core import IcaData
import pandas as pd
[8]:
new_sample_table = pd.DataFrame([['new_data','reference']]*2+[['new_data','test']]*2,columns=['project','condition'],index=log_tpm.columns)
new_sample_table
[8]:
| project | condition | |
|---|---|---|
| Reference_1 | new_data | reference |
| Reference_2 | new_data | reference |
| Test_1 | new_data | test |
| Test_2 | new_data | test |
[9]:
new_data = IcaData(ica_data.M,
activities,
gene_table = ica_data.gene_table,
sample_table = new_sample_table,
imodulon_table = ica_data.imodulon_table)
[10]:
from pymodulon.plotting import *
[11]:
plot_activities(new_data,'Fur-1',highlight='new_data')
[11]:
<AxesSubplot:ylabel='Fur-1 iModulon\nActivity'>

[ ]:
[ ]:
[ ]: